Code
Load packages
Customize ggplot themes
my_theme <- theme_bw() +
theme(
legend.position = "none",
axis.line.x = element_line(color = "black", size = 0.5),
axis.line.y = element_line(color = "white", size = 1),
axis.text.y = element_blank(),
axis.ticks = element_blank(),
panel.border = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank(),
panel.grid.major.y = element_blank())
my_theme2 <- theme_bw() +
theme(
legend.position = "none",
axis.line.x = element_line(color = "black", size = 0.5),
axis.line.y = element_line(color = "white", size = 1),
panel.border = element_blank(),
panel.grid.major.y = element_blank(),
panel.grid.minor.y = element_blank(),
axis.text.y = element_text(face = "italic"))
my_theme3 <- theme_bw() +
theme(
legend.position = "none",
axis.line.x = element_line(color = "black", size = 0.5),
axis.line.y = element_line(color = "white", size = 1),
panel.border = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.y = element_blank(),
legend.text = element_text(face = "italic"))Read data from XLSX
Summary and visualization
All strains per year
Fig1A <- fus %>%
tabyl(Host) %>%
ggplot(aes(reorder(Host, n), n, label = n))+
geom_col(fill = "#ffcc99", color = "black", size = 0.5)+
theme(axis.text.x=element_text(face="italic"))+
geom_text(position = position_dodge(width = 1),
hjust = 0.5, vjust = -0.2) +
labs(x = "", y = "Number of strains")+
scale_y_continuous(expand = c(0, 0), limits = c(0, 600)) +
my_themeAll strains per year
All strains per complexes
Fig1C <- fus %>%
tabyl(Complexes) %>%
ggplot(aes(reorder(Complexes, n),n, label = n))+
geom_col(fill = "#ffcc99", color = "black", size = 0.5)+
coord_flip()+
theme(axis.text.x=element_text(face="italic"))+
geom_text(position = position_dodge(width = 1),
hjust = 0) +
labs(x = "", y = "Number of strains")+
scale_y_continuous( limits = c(0, 650)) +
my_theme2Figure 1
fig1 <- ((Fig1A / Fig1B) | Fig1C) + plot_annotation(tag_levels = 'A')
ggsave("figs/fig1.png", width =6, height =4)
fig1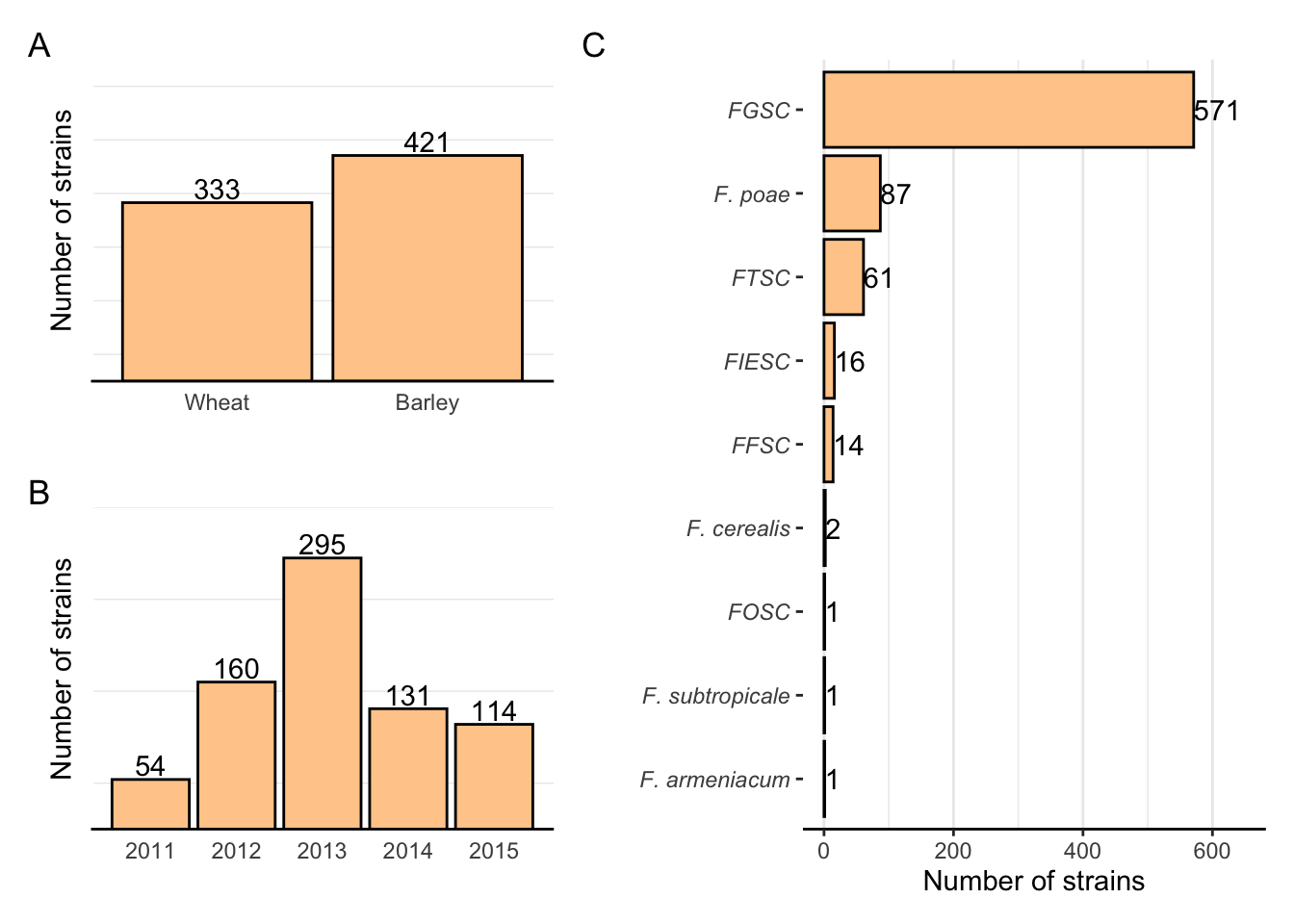
All species by host
Wheat
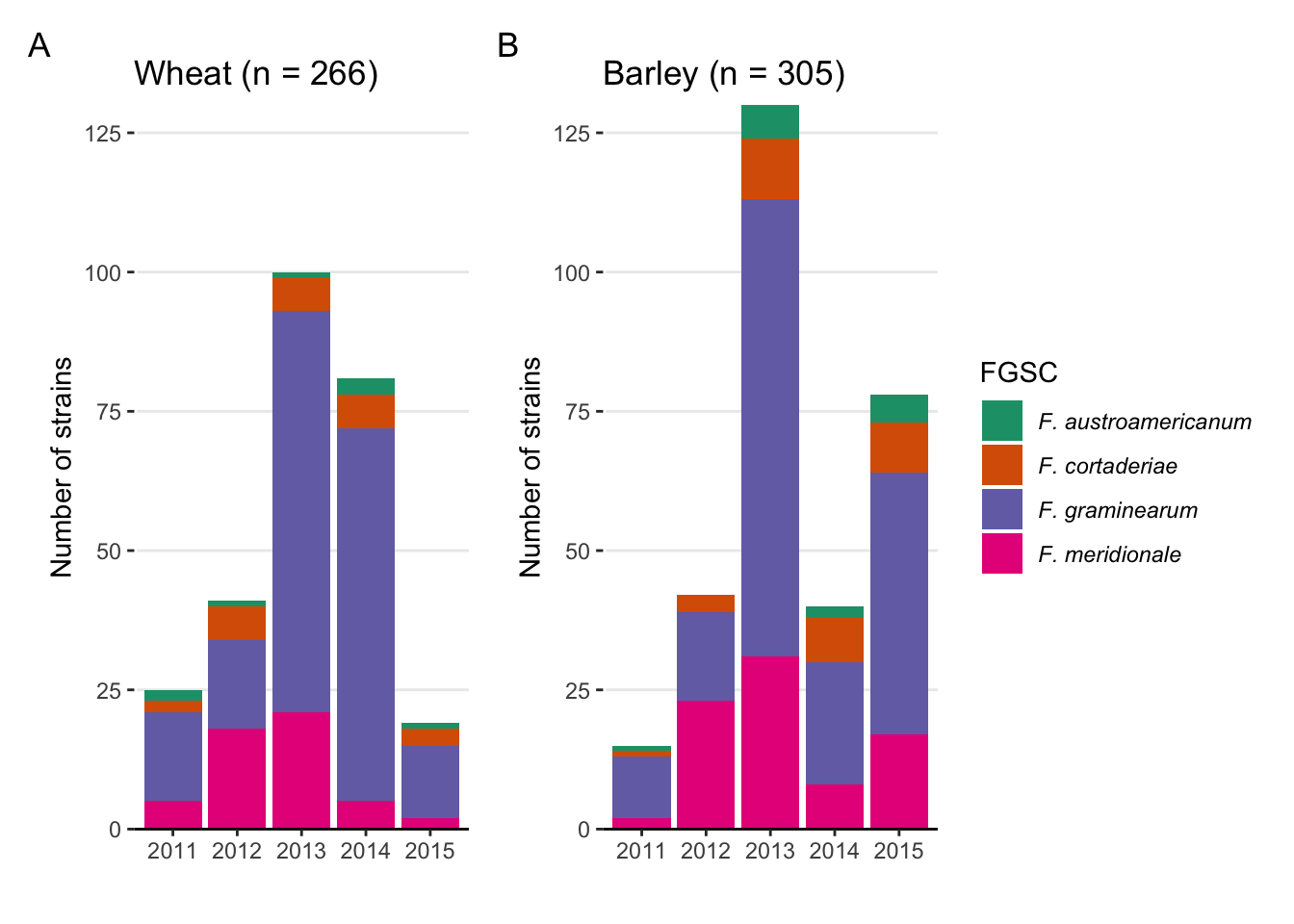
Fig2A <- fus %>%
filter(Host == "Wheat") %>%
tabyl(Complex2, Year) %>%
gather(year, n, 2:6) %>%
ggplot(aes(year, n, fill = Complex2))+
geom_col()+
scale_fill_brewer(palette = "Paired")+
theme_light()+
labs(x = "Year", y = "Number of strains", fill = "", title = "Wheat (n = 366)")+
scale_y_continuous(expand = c(0, 0), limits = c(0, 180)) +
my_theme3Barley
Fig2B <- fus %>%
filter(Host == "Barley") %>%
tabyl(Complex2, Year) %>%
gather(year, n, 2:6) %>%
ggplot(aes(year, n, fill = Complex2))+
geom_col()+
scale_fill_brewer(palette = "Paired")+
theme_light()+
labs(x = "Year", y = "Number of strains", fill = "", title = "Barley (n = 421)")+
scale_y_continuous(expand = c(0, 0), limits = c(0, 180)) +
my_theme3Figure 2
patch <- Fig2A + Fig2B & theme(legend.position = "right")
p1 <- patch + plot_layout(guides = "collect")+ plot_annotation(tag_levels = 'A')
ggsave("figs/fig2.png", width =7, height =4)
p1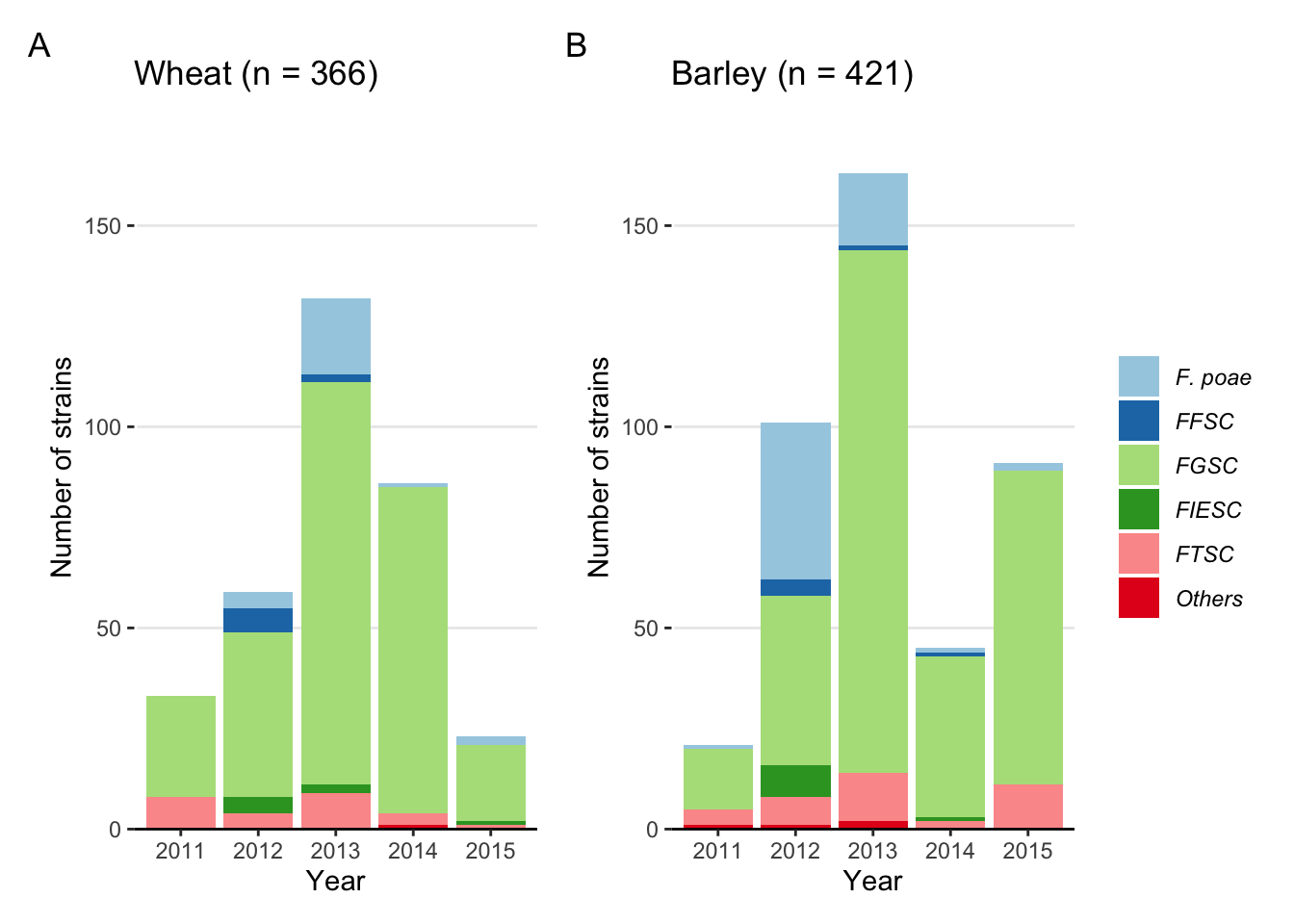
FGSC by host within year
Wheat
Fig3A <- fus %>%
filter(Complex2 == "FGSC") %>%
filter(Host == "Wheat") %>%
tabyl(Species, Year) %>%
gather(year, n, 2:6) %>%
ggplot(aes(year, n, fill = Species, label = n))+
geom_col()+
scale_fill_brewer(palette = "Dark2")+
theme_light()+
labs(x = "", y = "Number of strains", fill = "FGSC", title = "Wheat (n = 266)")+
scale_y_continuous(expand = c(0, 0), limits = c(0,130)) +
my_theme3Barley
Fig3B <- fus %>%
filter(Complex2 == "FGSC") %>%
filter(Host == "Barley") %>%
tabyl(Species, Year) %>%
gather(year, n, 2:6) %>%
ggplot(aes(year, n, fill = Species))+
geom_col()+
scale_fill_brewer(palette = "Dark2")+
theme_light()+
labs(x = "", y = "Number of strains", fill = "FGSC", title = "Barley (n = 305)")+
scale_y_continuous(expand = c(0, 0)) +
my_theme3FTSC by host within year
Wheat
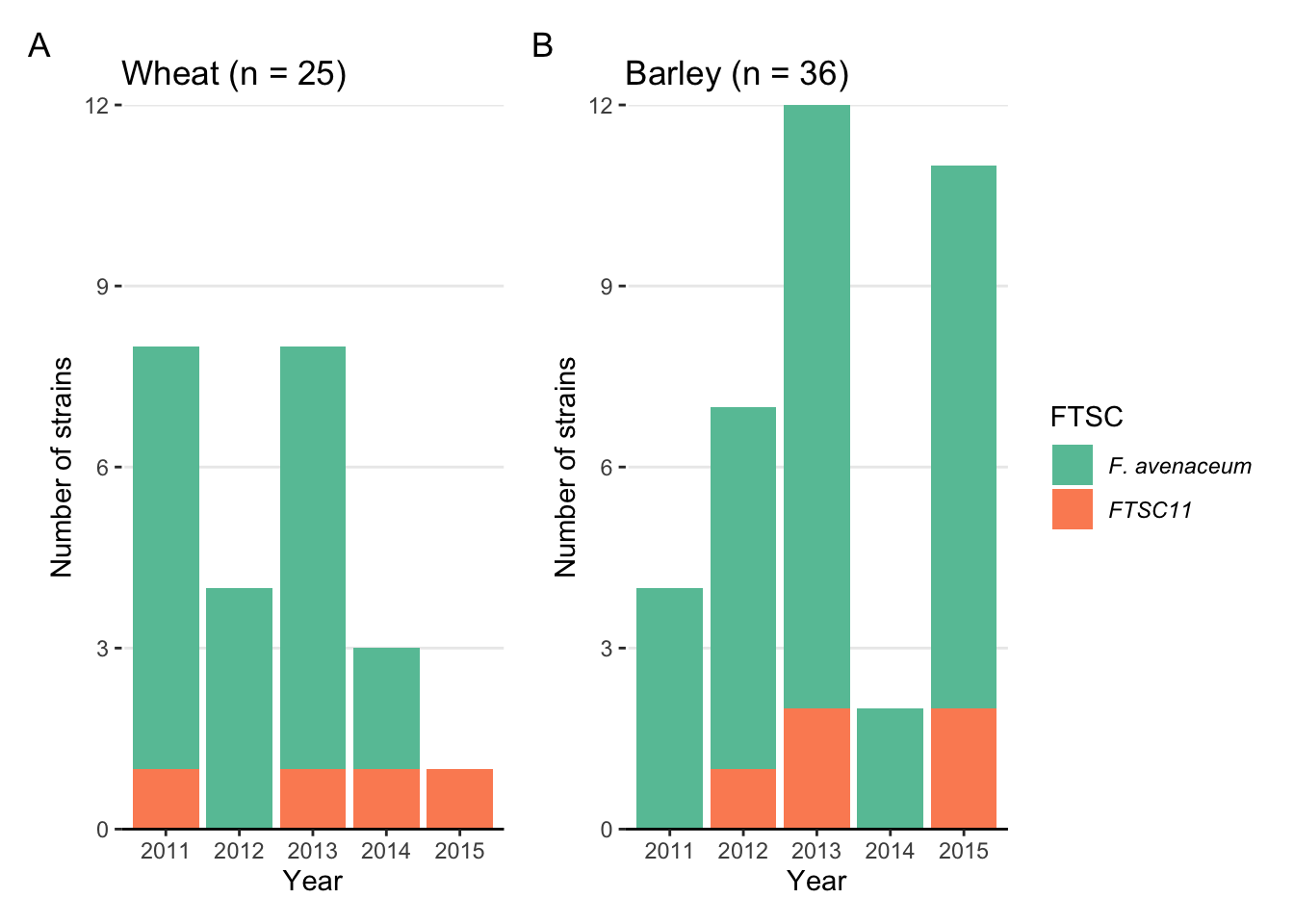
Fig3C <- fus %>%
filter(Complex2 == "FTSC") %>%
filter(Host == "Wheat") %>%
filter(Species != "F. acuminatum") %>%
tabyl(Species, Year) %>%
gather(year, n, 2:6) %>%
ggplot(aes(year, n, fill = Species))+
geom_col()+
scale_fill_brewer(palette = "Set2")+
theme_light()+
labs(x = "Year", y = "Number of strains", fill = "FTSC", title = "Wheat (n = 25)")+
scale_y_continuous(expand = c(0, 0), limits = c(0,12)) +
my_theme3Barley
Fig3D <- fus %>%
filter(Complex2 == "FTSC") %>%
filter(Host == "Barley") %>%
tabyl(Species, Year) %>%
gather(year, n, 2:6) %>%
ggplot(aes(year, n, fill = Species))+
geom_col()+
scale_fill_brewer(palette = "Set2")+
theme_light()+
labs(x = "Year", y = "Number of strains", fill = "FTSC", title = "Barley (n = 36)")+
scale_y_continuous(expand = c(0, 0), limits = c(0,12)) +
my_theme3Independence tests
Host vs. complexes
## Warning in stats::chisq.test(., ...): Chi-squared approximation may be incorrect##
## Pearson's Chi-squared test
##
## data: host_complexes
## X-squared = 12.159, df = 8, p-value = 0.1443##
## Fisher's Exact Test for Count Data
##
## data: host_complexes
## p-value = 0.07159
## alternative hypothesis: two.sidedyear vs. complexes
##
## Fisher's Exact Test for Count Data with simulated p-value (based on
## 2000 replicates)
##
## data: year_complex2
## p-value = 0.0004998
## alternative hypothesis: two.sidedFGSC vs. host
##
## Pearson's Chi-squared test
##
## data: host_fgsc
## X-squared = 7.3975, df = 3, p-value = 0.06025FGSC vs. year
year_fgsc <- fus %>%
filter(Complex2 == "FGSC") %>%
tabyl(Species, Year)
fisher.test(year_fgsc, simulate.p.value = T)##
## Fisher's Exact Test for Count Data with simulated p-value (based on
## 2000 replicates)
##
## data: year_fgsc
## p-value = 0.0004998
## alternative hypothesis: two.sided